About
Learn more about me
Computational & Evolutionary Biologist
- Website: taylorpaisie.github.io
- Phone: (419)672-1850
- City: Atlanta, GA
- Degree: MS
- Email: tpaisie91@gmail.com
I just really love infectious diseases of all kinds. Bacteria, viruses, all the good stuff. Computers are also a plus.
Skills
Interests
Phylodynamics
Bacerial and Viral Genomics
Animals
Computational Biology
Creating a sentient life from computer code
Senient life taking revenge on it's creators
CV
Taylor K. Paisie
Professional Experience
Bioinformatics Engineer, ORISE Fellow
2023 - Present
Atlanta, GA
Bioinformatics Engineer, Biotia, Inc.
2022 - 2023
Brooklyn, NY
Bioinformatics Scientist, University of Florida.
2022 - 2022
Gainesville, FL
Graduate Research Assistant, University of Florida.
2016 - 2021
Gainesville, FL
Education
PhD & Bioinformatics
2024 - Present
University of Cape Town, Cape Town, South Africa
Master of Science & Genetics and Genomics
2016 - 2021
University of Florida, Gainesville, FL
Bachelor of Science & Biology
2010 - 2014
Florida State University, Tallahasee, FL
Teaching Experience
Viral Evolution and Molecular Epidemiology (VEME) Workshop
August 4-9, 2024
Brasilia, Brazil
Viral Evolution and Molecular Epidemiology (VEME) Workshop
August 21-26, 2023
Stellenbosch, South Africa
Viral Evolution and Molecular Epidemiology (VEME) Workshop
August 21-26, 2022
Panama City, Panama
Data Carpentry Workshop: Introduction to Genomics
November 14-15, 2019
Gainesville, FL
Viral Evolution and Molecular Epidemiology (VEME) Workshop
August 4-9, 2019
Hong Kong, China
Data Carpentry Workshop: Introduction to Genomics
April 11-12, 2019
Gainesville, FL
Publications
Kim M, Enzler MJ, Pritt BS, Kumlien AC, Wolf MJ, Theel ES, Norgan AP, Paisie TK, Patel R (2024). The Brief Case: Human leptospirosis acquired in Minnesota and diagnosed using 16S ribosomal RNA gene PCR/next-generation sequencing of blood. Journal of Clinical Microbiology. https://doi.org/10.1128/jcm.00572-24.
Sabin SJ, Beesley CA, Marston CK, Paisie TK, Gulvik CA, Sprenger GA, Gee JE, Traxler RM, Bell ME, McQuiston JR, Weiner ZP (2024). Investigating Anthrax-Associated Virulence Genes Among Archival and Contemporary Bacillus cereus Group Genomes. Pathogens. https://doi.org/10.3390/pathogens13100884.
Alam MT, Stern SR, Frison D, Taylor K, Tagliamonte MS, Nazmus SS, Paisie TK, Hilliard NB, Jones RG, Iovine NM, Cherabuddi K, Mavian C, Myers P, Salemi M, Ali A and Morris JG (2023). Seafood-Associated Outbreak of ctx-Negative Vibrio mimicus Causing Cholera-Like Illness, Florida, USA. Emerging Infectious Diseases. . https://doi.org/10.3201/eid2910.230486.
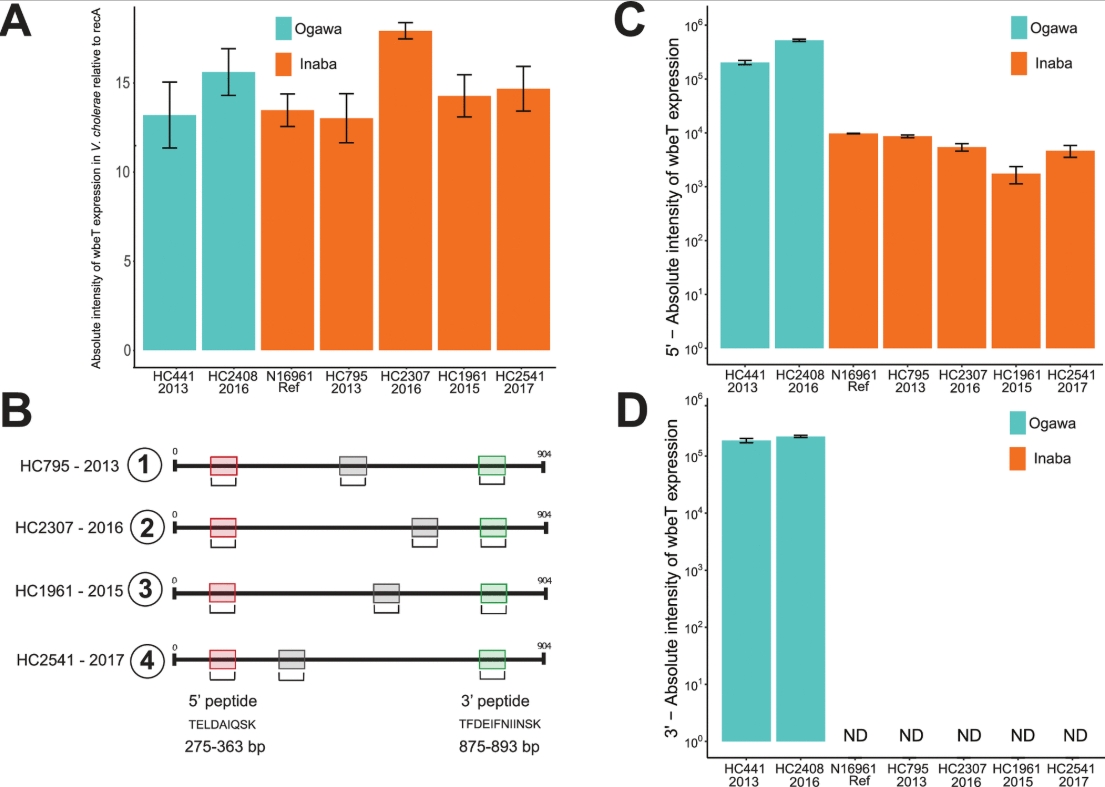
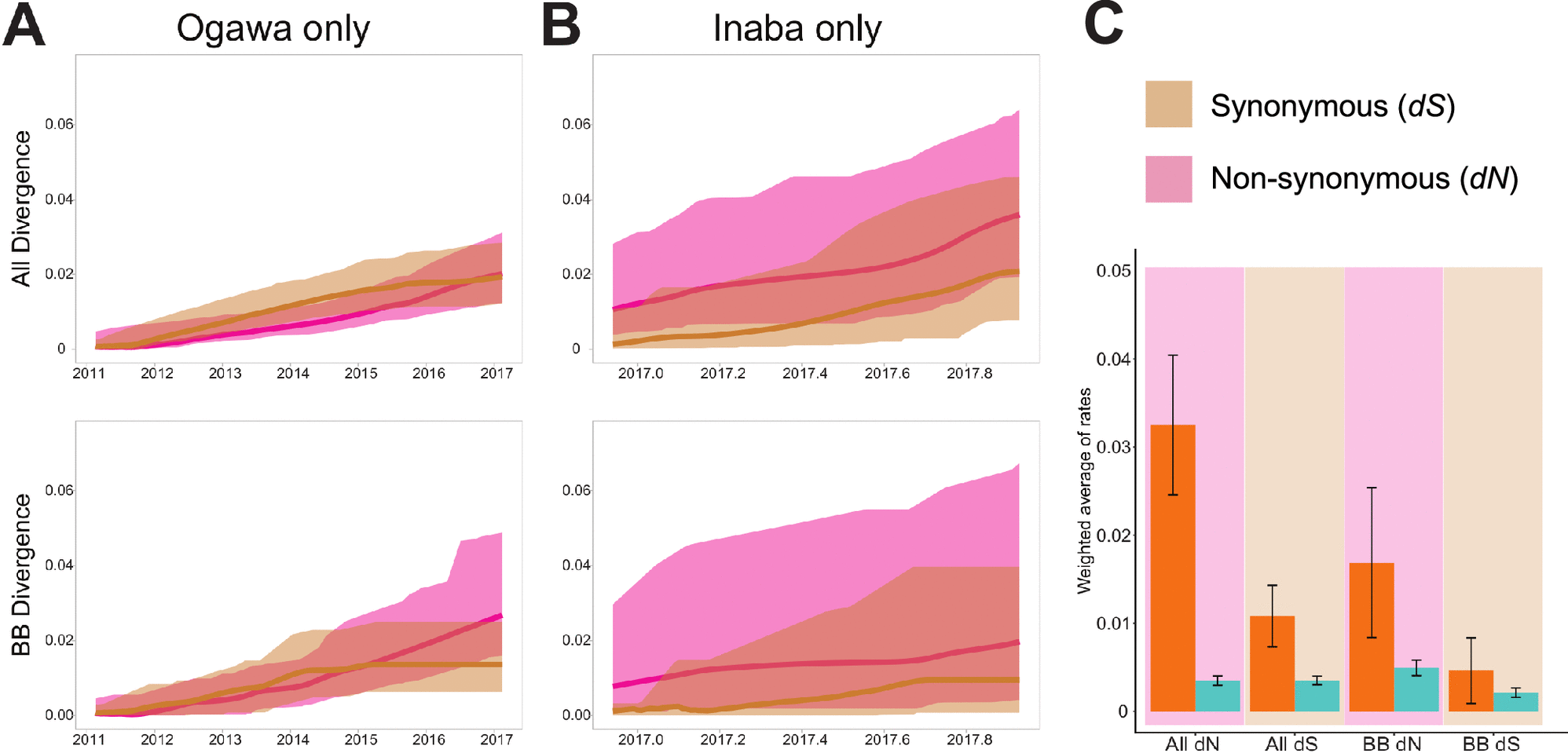
Paisie TK, Cash MN, Tagliamonte M, Ali A, Morris JG, Salemi M, Mavian C (2022). Molecular basis of the toxigenic Vibrio cholerae O1 serotype switch from Ogawa to Inaba in Haiti. Microbiology Spectrum. https://doi.org/10.1128/spectrum.03624-22.
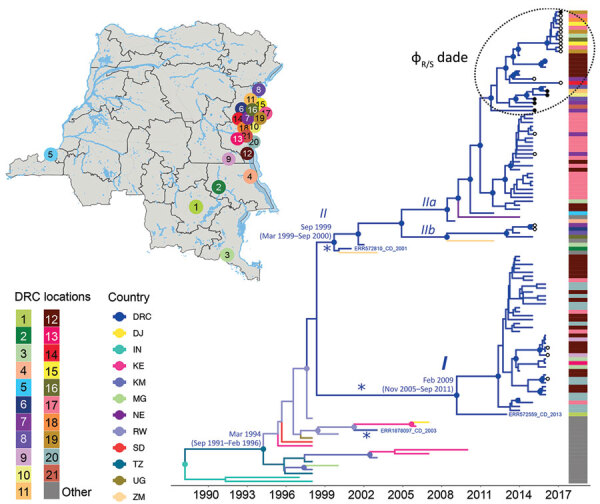
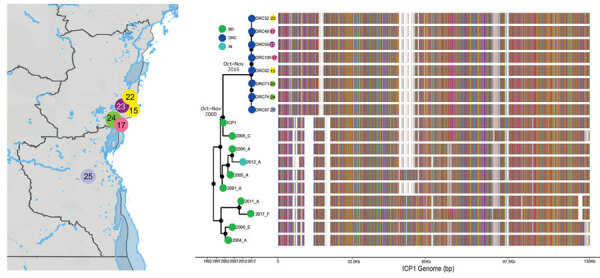
Alam MT, Mavian C, Paisie TK, Tagliamonte MS, Cash MN, Angermeyer A, Seed KD, Camilli A, Maisha FM, Kabangwa Kakongo Senga R, Salemi M, Morris JG, Ali A (2022). Emergence and Evolutionary Response of Vibrio cholerae to Novel Bacteriophage, Democratic Republic of the Congo. Emerging Infectious Diseases. https://doi.org/10.3201/eid2812.220572.
Sayeed MA, Paisie TK, Alam MT, Ali A, Camilli A, Wrammert J, Khan AI, Qadri F, Salemi M, Morris JG, Nelson EJ (2022). Development of a monoclonal antibody to a vibriophage as a proxy for Vibrio cholerae detection. Infection and Immunity. https://doi.org/10.1128/iai.00161-22. https://doi.org/10.1128/iai.00161-22.
Alexiev I, Mavian C, Paisie TK, Ciccozzi M, Dimitrova R, Gancheva A, Kostadinova A, Seguin-Devaux C, Salemi M (2022). Analysis of the Origin and Dissemination of HIV-1 Subtype C in Bulgaria. Viruses 2022, 14, 263. https://doi.org/10.3390/v14020263.
Mavian C, Tagliamonte M, Carreras BG, Bento, A, Paisie TK, Marini S, Cummings DA, Lednicky JA, Morris JG, Salemi M (2021). Multiple Introductions of Zika Virus in the Americas. Amer Soc Trop Med & Hygiene. Vol. 105. No. 5.
Mavian C, Pond SK, Marini S, Rife-Magalis B, Vandamme AM, Dellicour S, Samuel V. Scarpino, Houldcroft C, Villabona-Arenas J, Paisie TK, Trovao NS, Boucher C, Zhang Y, Scheuermann RH, Gascuel O, Lam TTY, Suchard MA, Abecasis A, Wilkinson E, de Oliveira T, Bento AI, Schmidt HA, Martin D, Hadfield J, Faria N, Grubaugh ND, Neher RA, Baele G, Lemey P, Stadler T, Albert J, Crandall KA, Leitner T, Stamatakis A, Prosperi M, Salemi M (2020). Sampling bias and incorrect rooting make phylogenetic network tracing of SARS-COV-2 infections unreliable. Proceedings of the National Academy of Sciences. https://10.1073/pnas.2007295117.
Mavian C*, Paisie TK*, Alam MT, Beau de Rochars VM, Nembrini S, Cash MN, Nelson EJ, Ali A, Morris JG, Salemi M (2020). Toxigenic Vibrio cholerae evolution and establishment of reservoirs in aquatic ecosystems. Proceedings of the National Academy of Sciences. https://10.1073/pnas.1918763117.
Ge Y, Paisie TK, Chen S, Concannon P (2019). UBASH3A regulates the synthesis and dynamics of T-cell receptor-CD3 complexes. The Journal of Immunology. https://doi:10.4049/jimmunol.1801338.
White SK, Mavian C, Elbadry MA, Beau de Rochars VM, Paisie T, Telisma T, Salemi M, Lednicky JA, Morris JG (2018). Detection and phylogenetic characterization of arbovirus dual-infections among persons during a Chikungunya Fever outbreak, Haiti 2014. PLoS Negl Trop Dis 12(5): e0006505. https://doi.org/10.1371/journal.pntd.0006505.
Blohm G, Lednicky JA, Marquez M, White SK, Loeb JC, Pacheco C, Nolan DJ, Paisie T, Salemi M, Rodriguez-Morales A, Morris JG, Pulliam J, Carrillo A, Plaza J, Paniz-Mondolfi A (2017). Evidence for mother-to-child transmission of Zika virus through breast milk. Clinical Infectious Diseases. https://doi.org/10.1093/cid/cix968.
Ge, Y, Paisie TK, Newman, JRB, McIntyre, LM, Concannon, P (2017). UBASH3A Mediates Risk for Type 1 Diabetes through Inhibition of T-Cell Receptor-Induced NF-B Signaling. Diabetes. https://doi.org/10.2337/db16-1023.
Blohm G, Lednicky JA, Marquez M, White SK, Loeb JC, Pacheco C, Nolan DJ, Paisie T, Salemi M, Rodriguez-Morales A, Morris JG, Pulliam J, Carrillo A, Plaza J, Paniz-Mondolfi A (2017). Complete Genome Sequences of Identical Zika Viruses in a Nursing Mother and Her Infant. Genome Announc 5:e00231-17. https://doi.org/10.1128/genomeA.00231-17.
Bonny TS, Driver JP, Paisie T, Salemi M, Morris JG, Shender LA, Smith L, Enloe C, Oxenrider K, Gore JA, Loeb JC, Wu C-Y, Lednicky JA. Detection of Alphacoronavirus vRNA in the Feces of Brazilian Free-Tailed Bats (Tadarida brasiliensis) from a Colony in Florida, USA (2017). Diseases. 10.3390/diseases5010007.
Cherabuddi K, Iovine, NM, Shah K, White SK, Paisie T, Salemi M, Lednicky, J (2016). Zika and chikungunya virus co-infection in a traveller returning from Colombia: virus isolation and genetic analysis. JMM Case Reports. http://doi.org/10.1099/jmmcr.0.005072.
Mason OU, Canter E, Gillies LE, Paisie T, and Roberts BJ (2016). Mississippi River plume enriches microbial diversity in the northern Gulf of Mexico. Front. Microbiol. 7:1048. doi: 10.3389/fmicb.2016.01048.
Paisie TK, Miller TE, Mason OU (2014) Effects of a Ciliate Protozoa Predator on Microbial Communities in Pitcher Plant (Sarracenia purpurea) Leaves. PLoS ONE 9(11): e113384. doi:10.1371/journal.pone.0113384.
Portfolio
My Works
Data Visualization Dashboards
Assembly Metrics Visualization Dashboard
Assembly Metrics Visualization Dashboard Github
SNP Workflow Visualization Dashboard
SNP Workflow Visualization Dashboard Github
Phylogenetic Tree and Map Visualization Dashboard
Phylogenetic Tree and Map Visualization Dashboard Github
Tutorials and Lessons
- All
Contact
Contact Me

Computational & Evolutionary Biologist
- Birthday: 25 May 1991
- Website: taylorpaisie.github.io
- Phone: (419)672-1850
- City: Atlanta, GA
- Age: 33
- Degree: MS
- Email: tpaisie91@gmail.com
I just really love infectious diseases of all kinds. Bacteria, viruses, all the good stuff. Computers are also a plus.
Social Profiles
Email Me
tpaisie91@gmail.com
Call Me
(419)672-1850